Recently, I migrated our AWS Glue Catalog permissions to be managed with AWS Lake Formation. AWS Lake Formation adds a layer of security which allows fine-grained permissions, manage data lake resource locations (s3) as well as control permissions by Tags (LF-Tag) and column level permission management to IAM Users, Roles, Cross-Account and SAML or QuickSight User ARN. AWS Lake Formation was added after AWS Glue Catalog service, and in order to continue supporting backward compatibility, Lake Formation by default Grants IAMAllowedPrincipals (basically, doesn’t evaluate permissions) access to Glue Catalog resources as long as Principal has appropriate IAM Policy.
Setup
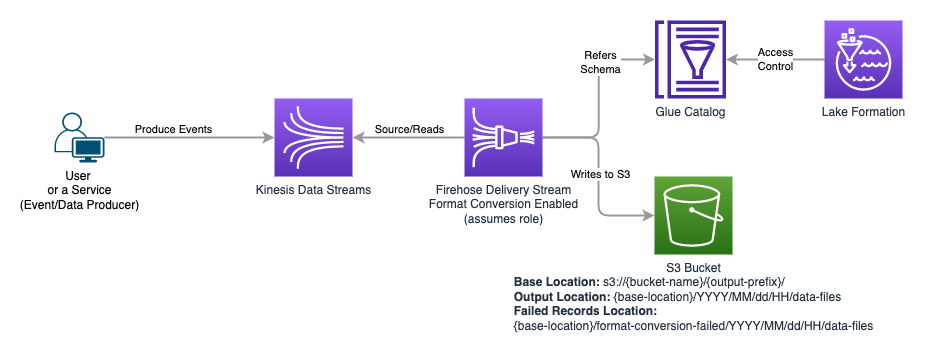
Set of services which produce events/messages and send to Kinesis Data Streams, which is read by Firehose Delivery Stream with Record Format Conversion Enabled to convert JSON format data from Kinesis Stream to Parquet (Columnar) format with compression so that querying that data using Athena (and other tools) is optimized for Cost and Speed. In this setup, Database, Table and Schema is configured in Glue Catalog, which is referred by Firehose to convert JSON to Parquet.
What we missed
We did take care of granting appropriate permissions to Users and Roles which users assume, however, we had few AWS Kinesis Data Firehose streams in place, which had Data Format conversion enabled, to convert JSON into Parquet, referring to Schema and Table registered in Glue Catalog. Unfortunately, we forgot to include roles assumed by Firehose in Lake Formation permission. Once we realized that the Firehose was failing to write files to S3 as it could no longer read Glue Table from Catalog, we went ahead and fixed the issue by granting SELECT permission on Glue Table to roles assumed by Firehose.
With this incident, we had gap of data in S3. As Firehose works on streaming (batches of 5–15 minutes or size of the batch) basis, during the window when Firehose role didn’t have permission to Glue Table, the data it failed to write due to this permission problem was not available in parquet converted output location. For any legitimate Format Conversion issues, firehose writes failed record to sub-folder in S3, unless you override, it is in format-conversion-failed folder of your target S3 location (bucket + prefix).
Data (with failed records) in S3
S3 has parquet converted data regularly flowing in, however, there is gap in and between highlighted timeframe of 2021/11/04/02 and 2021/11/06/04.
Data now showed up as parquet in parquet converted output location, we find that those files as raw data (json) exists in format-conversion-failed subdirectory (prefix) in S3.
Solution
We have raw data in format-conversion-failed subdirectory, and we need to convert that to parquet and put it under parquet output directory, so that we fill the gap caused by permission issue, which lasted for about two days.
- Read Glue schema, which is also used by Firehose for format conversion
- Read one file at a time from
format-conversion-failedsubdirectory for the date/time range when we saw the issue - Convert each JSON to parquet, with schema downloaded from step 1
- Write parquet file to respective S3 parquet output directory
Download failed JSON files from S3
# date when issue started - just for local storage
ISSUE_DATE=20211104
ERROR_PREFIX=format-conversion-failed# Create local directory - change `/tmp` prefix as appropriate
mkdir -p "/tmp/${ISSUE_DATE}/${FIREHOSE_OUTPUT_PATH}"
cd "/tmp/${ISSUE_DATE}/${FIREHOSE_OUTPUT_PATH}"# Download failed files from S3 to local
aws s3 --profile ${aws-profile} cp --recursive "s3://${BUCKET}/${FIREHOSE_OUTPUT_PATH}/${ERROR_PREFIX}/" "./${ERROR_PREFIX}"# Count all downloaded failed files
find "./${ERROR_PREFIX}" -type f | wc -l
Extract rawData from failed JSON files
Bash to Convert errored files from format-conversion-failed errored object with rawData (which is base64 encoded), and doing base64 decode and writing to “./json” directory.
mkdir json# Loop through each JSON file, grab rawData fields from each object, and perform base64 decode and write to a separate JSON file
for f in $(find ./${ERROR_PREFIX} -type f | sort); do
json=$(echo $f | sed "s/format-conversion-failed/json/g");
dir=$(echo "${json%/*}"); mkdir -p $dir;
for r in $(cat $f | jq '.rawData' -r); do echo $r | base64 -d; done > $json
echo $f:$(cat $f | wc -l):$json:$(cat $json | wc -l) | tee -a /tmp/failed-to-json-counts
done# Count number of files with actual JSON extracted (and base64 decoded) from rawData field
find "./json" -type f | sort | wc -l
Prepare JSON file list for Spark
From ./json directory, list all files and sort it by name (which includes path), and write it to a file. (Note, we could perform equivalent step in spark as well)
find ./json -type f | sort > files.txt
Download Glue Schema used by Firehose
Copy JSON format Avro schema from Glue Catalog which is used by Firehose to convert JSON to Parquet, to a schema file, schema.json
Spark Code to convert JSON to Parquet, using schema.json
Reading data from ./json to ./parquet with parquet files, by applying schema.json
Start Spark Shell
spark-shell — packages org.apache.spark:spark-avro_2.12:3.1.1
Spark Code
import spark.implicits._
import org.apache.spark.sql.functions.unbase64
import java.io._
import java.nio.file._
import org.apache.spark.sql.avro.functions._
import org.apache.spark.sql.types._
import org.apache.spark.sql.avro._
import scala.reflect.io.Directorydef getListOfFiles(dir: File, extensions: List[String]): List[File] = {
dir.listFiles.filter(_.isFile).toList.filter { file =>
extensions.exists(file.getName.endsWith(_))
}
}val SCHEMA_FILE = "{path-to-schema.json}"
val jsonSchema = new String(Files.readAllBytes(Paths.get(SCHEMA_FILE)))
val schema = spark.read.format("avro").option("avroSchema", jsonSchema).load().schemaval filesDF = spark.read.text("files.txt")
filesDF.show
filesDF.countfilesDF.take(filesDF.count.asInstanceOf[Int]).foreach {
row => {
//row.toSeq.foreach{col => println(col + " = " + col.asInstanceOf[String])}
val jsonFile = row.get(0).asInstanceOf[String]
val parquetDir = jsonFile.replace("./json/", "./parquet/")
println(jsonFile + " = " + parquetDir)
val df = spark.read.schema(schema).json(jsonFile)
val numRecords = df.count()df.coalesce(1).write.option("compression", "snappy").mode("overwrite").parquet(parquetDir)
val parquetFile = getListOfFiles(new File(parquetDir), List("parquet"))(0)
println("Source: " + jsonFile + ", outputFile: " + parquetFile.toPath + " renamed to " + parquetDir + ".parquet" + ", numRecords: " + numRecords)
Files.move(parquetFile.toPath, new File(parquetDir + ".parquet").toPath, StandardCopyOption.ATOMIC_MOVE)
new Directory(new File(parquetDir)).deleteRecursively()
}
}
Count number of Parquet files
find ./parquet -type f | sort | wc -l
Validate Counts in JSON and Parquet files
# Count of records per JSON file
for f in $(find ./json -type f | sort); do echo $f: $(cat $f | wc -l); done > /tmp/json.counts # Count of records per Parquet file
for f in $(find ./parquet -type f | sort); do echo $f: $(parquet-tools rowcount $f | awk -F':' '{print $2}'); done | sed 's/parquet\//json\//g' | sed 's/.parquet//g' > /tmp/parquet.counts# Compare two files, each having counts from JSON and Parquet file
# Both files must have matching number of JSON and Parquet records
diff /tmp/json.counts /tmp/parquet.counts
Upload Parquet files to S3
# Dry run to upload Parquet files
aws s3 --profile ${aws-profile} cp --recursive --dryrun parquet/ "s3://${BUCKET}/${FIREHOSE_OUTPUT_PATH}/"# Upload files after verifying Dry Run output
aws s3 --profile ${aws-profile} cp --recursive parquet/ "s3://${BUCKET}/${FIREHOSE_OUTPUT_PATH}/"
After applying all steps above, gap in the files in S3 for parquet output should no longer be there.
Note that it is possible to convert bash script steps to Spark. I have just outlined the steps I followed to resolve the issue of missing data. When I consolidate these steps into a single Spark script, I will update it here. If you have already solved this, please let me know and I can reflect it here.
As always, please share if you have encountered such issues and have come up with better ways to resolve.