Access Cancer Database TCGA By R Code
Introduction
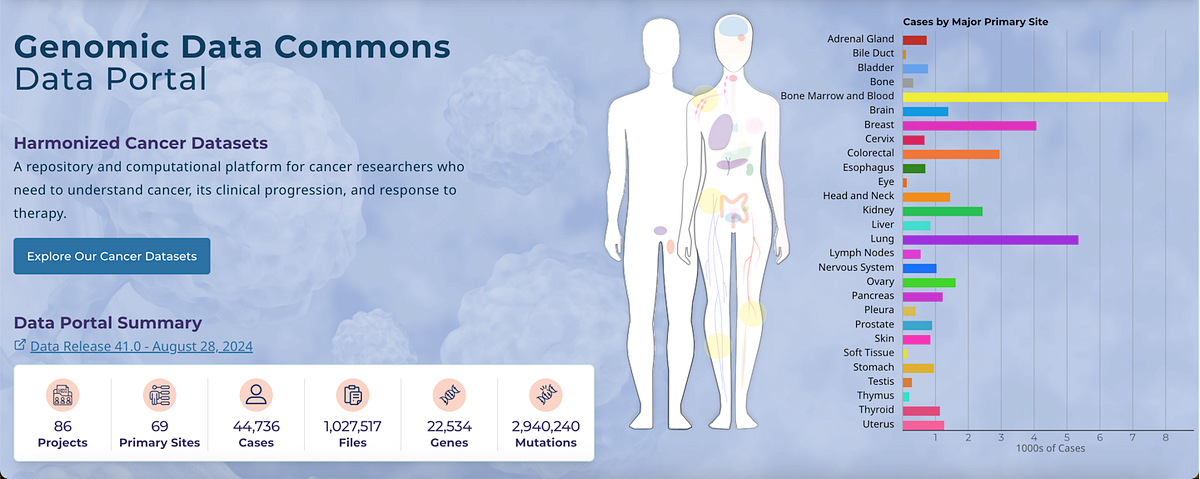
The Cancer Genome Atlas (TCGA), is a project funded by the National Institutes of Health (NIH), to map the genetic mutations that are responsible for cancer. TCGA collected a wide range of datasets of genomics, transcriptomics, epigenomics, and proteomics data with 33 types of cancer from many cancer patients.
The data is publicly available by multiple platforms, like Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov/), and can also be accessed programmatically via R tools by using Bioconductor packages. This way it will make it easier for researchers to download and process large amounts of datasets for cancer research.
The example code for downloading brain cancer data from TCGA using R, first needs to upload all the required packages and libraries.
Steps
Here I am providing steps by steps guide to download the big data from TCGA of different cancers by using simple code in R.
1. Install Necessary Packages
First, we need to install the TCGAbiolinks, is a R/Bioconductor package that helps to access, download, and retrieve large amounts of cancer-related data from…
Learn more How to Download and Manage Cancer Data From TCGA Using R